Welcome to the CBIO website
The Centre for Computational Biology is a Common Research Centre of MINES ParisTech, one of the major engineering schools for applied mathematics in France, and ARMINES, a private non-profit research and technological organisation. Our research is dedicated to the development of methods and tools in the fields of Machine Learning, Statistics and Computer Vision in order to analyze massive data generated in life sciences and medicine. We work on a broad range of applications, from questions in fundamental life science to precision medicine. The Centre for Computational Biology has a partnership with Inserm, the French national institute of health and medical research, and Institut Curie, a major hospital and research center dedicated to cancer. This partnership provides access to the infrastructure and facilities of the Institut Curie and facilitates collaborative projects with other groups at the Institut Curie, as well as data sharing. Our laboratory is located in the heart of Paris and we therefore benefit from an exceptional scientific and cultural environment.
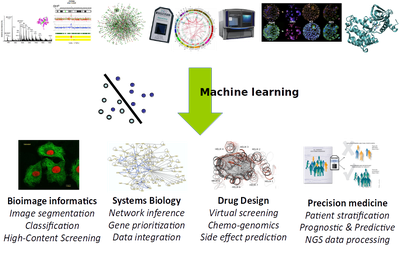
Keywords: computational biology, bioinformatics, machine learning, statistics, precision medicine, bioimaging, bioimage informatics, computer vision, virtual screening, chemoinformatics, genetics, systems biology, big data
